Fun case of speeding up data retrieval from PostgreSQL with Psycopg en ru
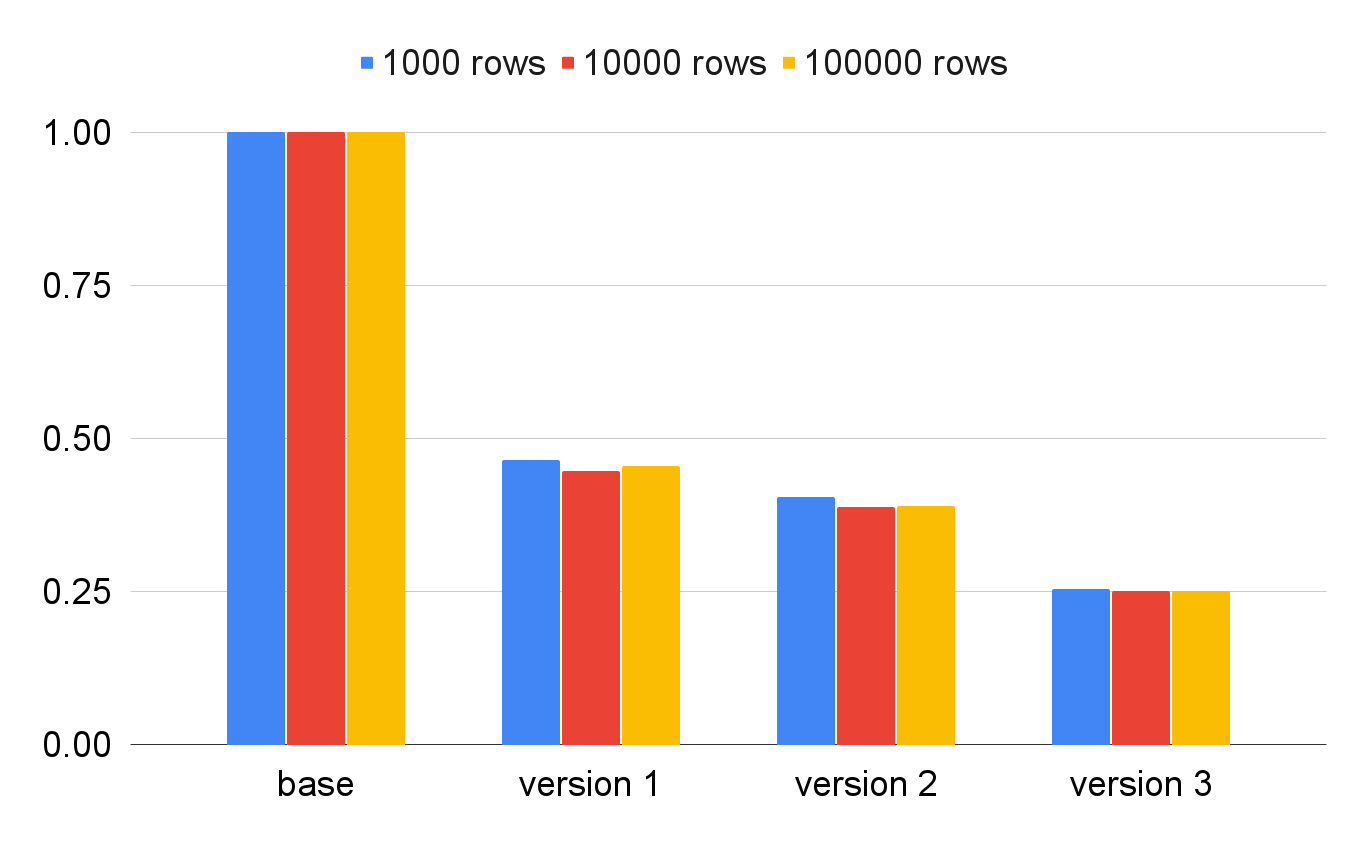
Speed of data retrieval from the PostgreSQL for each of the optimizations. In percentages of the base implementation speed. Note how the number of retrieved rows has little effect on execution time.
Once in a year or two, I have to remember that Python, umm... is not C++. It usually happens suddenly, like this time.
After a thoughtful analysis I got tired of waiting 10 seconds for news to load on feeds.fun, so I rolled up my sleeves and started optimizing. I almost jumped into an epic refactoring, but remembered just in time: measure first, then cut. In this case, the advice is literal — I took a profiler — py-spy — and checked what exactly was causing the slowdown
It turned out that the problem is not in the entire logic, but in a very specific place where the code extracts ~100000 rows from a PostgreSQL table, plus-minus 20000. The indexes are in place, I ran the tests with a database on a RAM disk, so everything should have been fine from the database side.
Don't be surprised by such a large number of rows:
- Firstly, I have a substantial news flow.
- Secondly, the reader currently assigns about 100 tags for each news item.
Armed with py-spy and the sources of psycopg, I went through three stages of optimization, reducing the function execution time approximately 4x solely by changing the format of the requested columns in SELECT query and the result processing code.
In the following text, I will tell you about the sequence of curious discoveries I made in the process.
Attention!
This post is not a study of Psycopg or Python performance, but a description of a specific experience with a specific task and specific data.
It would be incorrect to judge the performance of Psycopg, or Python in general, based on a single specific case.
Original task
Here is a shortened description of the table where the data is stored (I removed the extra to avoid formatting issues):
ffun=# \d o_relations
Table "public.o_relations"
Column | Type | Collation | Nullable |
------------+--------------------------+-----------+----------+
id | bigint | | not null |
entry_id | uuid | | not null |
tag_id | bigint | | not null |
created_at | timestamp with time zone | | not null |
Indexes:
"o_relations_pkey" PRIMARY KEY, btree (id)
"idx_o_relations_entry_id_tag_id" UNIQUE, btree (entry_id, tag_id)
The task of the target Python function: extract all tag_id for the list of entry_id passed and return a dictionary with a set of tag_id for each entry_id.
The result of the function should look like this:
{
"uuid-1": {1, 2, 3, 4, 5},
"uuid-2": {1, 7, 8},
....
}
No magic, just one SELECT plus dictionary creation.
Caveats and test description
To avoid the influence of external factors, I slightly simplified the original task:
- Instead of passing a list of
entry_id, I passed the number of records to extract (1000, 10000, 100000). - I did not use the Psycopg dict_row factory to exclude unnecessary data transformations.
- the measured functions are synchronous, whereas the original code was asynchronous.
- The test data was taken from production.
Also, note:
- I measured the execution time of the target Python function with all necessary data transformations, not just the Psycopg execution time. This is because the focus is on the speed of achieving the desired result, not merely retrieving "any" data from the database.
- I tried explicitly enabling the binary communication protocol with PostgreSQL, but the changes were negligible, so I don't mention this option further.
- Before measuring, each measured function was executed once to warm up the database.
- To measure each function, I made 100 runs and averaged the results.
The average execution time of the function was:
- For the base version: 2.28, 23.18, 227.91 seconds for 1000, 10000, 100000 records, respectively.
- For the final version: 0.58, 5.83, 57.27 seconds for 1000, 10000, 100000 records, respectively.
But keep in mind, the task and data are specific to a particular project, and even to my specific account within it, so they may not be very meaningful to an outside reader.
The complete test code
import uuid
import psycopg
import timeit
N = 1000
M = 100
def version_1(conn, n):
data = {}
with conn.cursor() as cur:
cur.execute("SELECT * FROM o_relations ORDER BY id DESC LIMIT %(limit)s", {"limit": n})
results = cur.fetchall()
for row in results:
entry_id = row[1]
if entry_id not in data:
data[entry_id] = set()
data[entry_id].add(row[2])
return data
def version_2(conn, n):
data = {}
with conn.cursor() as cur:
cur.execute("SELECT entry_id, tag_id FROM o_relations ORDER BY id DESC LIMIT %(limit)s", {"limit": n})
results = cur.fetchall()
for row in results:
entry_id = row[0]
if entry_id not in data:
data[entry_id] = set()
data[entry_id].add(row[1])
return data
def version_3(conn, n):
data = {}
entry_ids_mapping = {}
with conn.cursor() as cur:
cur.execute(
"SELECT entry_id::text, tag_id FROM o_relations ORDER BY id DESC LIMIT %(limit)s", {"limit": n}
)
results = cur.fetchall()
for row in results:
raw_entry_id = row[0]
if raw_entry_id not in entry_ids_mapping:
entry_ids_mapping[raw_entry_id] = uuid.UUID(raw_entry_id)
entry_id = entry_ids_mapping[raw_entry_id]
if entry_id not in data:
data[entry_id] = set()
data[entry_id].add(row[1])
return data
def version_4(conn, n):
data = {}
entry_ids_mapping = {}
with conn.cursor() as cur:
cur.execute(
"SELECT CONCAT(entry_id::text, '|', tag_id::text) AS ids FROM o_relations ORDER BY id DESC LIMIT %(limit)s",
{"limit": n},
)
results = cur.fetchall()
for row in results:
raw_entry_id, raw_tag_id = row[0].split("|")
if raw_entry_id not in entry_ids_mapping:
entry_ids_mapping[raw_entry_id] = uuid.UUID(raw_entry_id)
entry_id = entry_ids_mapping[raw_entry_id]
if entry_id not in data:
data[entry_id] = set()
data[entry_id].add(int(raw_tag_id))
return data
def run():
with psycopg.connect("dbname=ffun user=ffun password=ffun host=postgresql") as conn:
##########################################################################
# Demonstrate that everything works correctly and the results are the same
# Also, warm up the database
##########################################################################
data_1 = version_1(conn, N)
data_2 = version_2(conn, N)
data_3 = version_3(conn, N)
data_4 = version_4(conn, N)
print(data_1 == data_2 == data_3 == data_4)
##########################################################################
# Measure the performance of each version
##########################################################################
time_1 = timeit.timeit(lambda: version_1(conn, N), number=M)
time_2 = timeit.timeit(lambda: version_2(conn, N), number=M)
time_3 = timeit.timeit(lambda: version_3(conn, N), number=M)
time_4 = timeit.timeit(lambda: version_4(conn, N), number=M)
print(f"Number of records: {N}")
print(f"Number of measurements: {M}")
print(f"Version 1: {time_1:.2f} seconds")
print(f"Version 2: {time_2:.2f} seconds")
print(f"Version 3: {time_3:.2f} seconds")
print(f"Version 4: {time_4:.2f} seconds")
run()
The base version of the measured function:
def version_1(conn, n):
data = {}
with conn.cursor() as cur:
cur.execute("SELECT * FROM o_relations ORDER BY id DESC LIMIT %(limit)s", {"limit": n})
results = cur.fetchall()
for row in results:
entry_id = row[1]
if entry_id not in data:
data[entry_id] = set()
data[entry_id].add(row[2])
return data
Optimization 1
The first thing the profiler showed was a lot of time spent in psycopg/types/datetime.py — more than 18%! As you may have noticed, there is no time-related work in the function code.
"Ah-ha," I said to myself, "You, Tiendil, put a star in the SELECT, but you only need two columns, and parsing time values tends to be expensive."
And I replaced the star with specific columns:
def version_2(conn, n):
data = {}
with conn.cursor() as cur:
cur.execute("SELECT entry_id, tag_id FROM o_relations ORDER BY id DESC LIMIT %(limit)s", {"limit": n})
results = cur.fetchall()
for row in results:
entry_id = row[0]
if entry_id not in data:
data[entry_id] = set()
data[entry_id].add(row[1])
return data
Optimization 2
The next profiler run showed that times improved, but much time was still spent parsing the UUID values — this is the type of the entry_id column.
As I mentioned earlier, the peculiarity of the data is that for a single entry_id, there can be around 100 rows in the table. Therefore, it makes no sense to parse entry_id for each row.
What if we retrieve entry_id as a string and parse it on the Python side, but only once for each unique value?
def version_3(conn, n):
data = {}
entry_ids_mapping = {}
with conn.cursor() as cur:
cur.execute(
"SELECT entry_id::text, tag_id FROM o_relations ORDER BY id DESC LIMIT %(limit)s", {"limit": n}
)
results = cur.fetchall()
for row in results:
raw_entry_id = row[0]
if raw_entry_id not in entry_ids_mapping:
entry_ids_mapping[raw_entry_id] = uuid.UUID(raw_entry_id)
entry_id = entry_ids_mapping[raw_entry_id]
if entry_id not in data:
data[entry_id] = set()
data[entry_id].add(row[1])
return data
Optimization 3
It got even better (see the cover image).
This time py-spy led me to a much more interesting place: psycopg/pq/pq_ctypes.py, or more precisely to PGresult.get_value.
PGresult.get_value returns a single value from the query result by numbers of row and column. Along the way, it converts data from C format to Python format, in particular, using ctypes.string_at.
Data conversion from C to Python is a very costly operation. Actually, scratch that — it's a VERY costly operation. Especially when there are many such conversions, for instance, two for each of 100000 rows.
Is it possible to reduce the number of conversions? Of course, let's concatenate the columns on the database side and split them back on the Python side.
And here is our final version:
def version_4(conn, n):
data = {}
entry_ids_mapping = {}
with conn.cursor() as cur:
cur.execute(
"SELECT CONCAT(entry_id::text, '|', tag_id::text) AS ids FROM o_relations ORDER BY id DESC LIMIT %(limit)s",
{"limit": n},
)
results = cur.fetchall()
for row in results:
raw_entry_id, raw_tag_id = row[0].split("|")
if raw_entry_id not in entry_ids_mapping:
entry_ids_mapping[raw_entry_id] = uuid.UUID(raw_entry_id)
entry_id = entry_ids_mapping[raw_entry_id]
if entry_id not in data:
data[entry_id] = set()
data[entry_id].add(int(raw_tag_id))
return data
The performance of this version is approximately 4 times faster than the baseline (on my dataset).
Just to spell it out clearly
Formatting and concatenating result columns on the PostgreSQL side and then parsing them in Python might be faster than fetching the columns as separate values.
At least, this holds true for my specific use case with Psycopg. I love this library and believe in its quality, so I suspect the alternatives wouldn't fare any better.
Read next
- Notes on backend metrics in 2024
- Dungeon generation — from simple to complex
- Generation of non-linear quests
- Top LLM frameworks may not be as reliable as you think
- Prompt engineering: building prompts from business cases
- Two years of writing RFCs — statistics
- World Builders 2023: Preferences of strategy players
- World Builders 2023: Preparing a business plan for a game on Steam
- Pricing model at the start of Feeds Fun monetization
- Feeds Fun: marketing test — or how I blew 650 euros